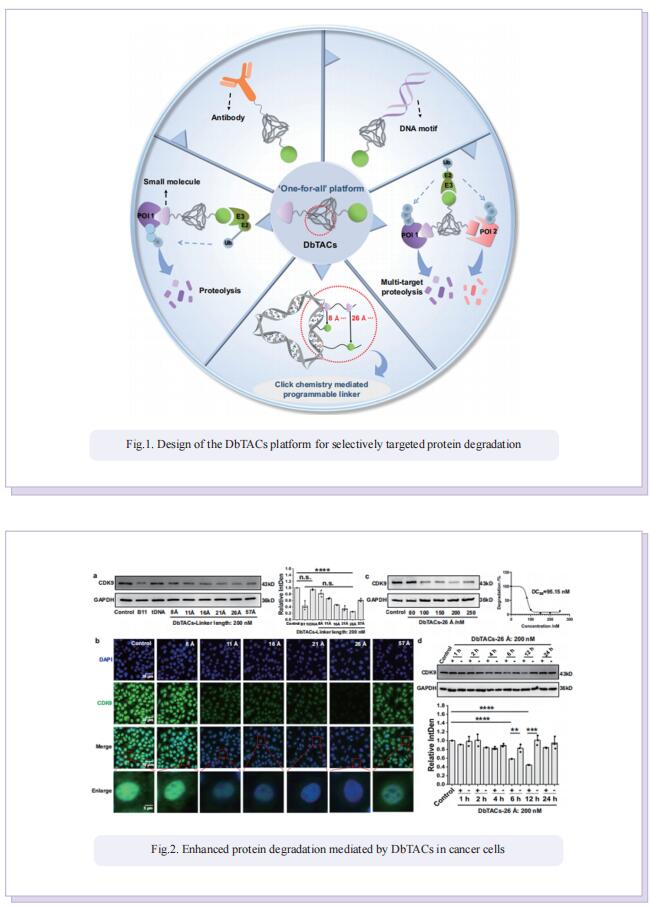
DNA framework-engineered chimeras platform enables selectively targeted protein degradation
Protein degradation is an emerging strategy to treat diseases, especially proteolysis targeting chimeras (PROTACs). PROTACs are heterobifunctional molecules that comprise a ligand targeting the protein of interest (POI), an element that recruits E3 ligases, and a linker connecting the above two moieties. Among them, the linker plays a critical role in bridging these two moieties. While many traditional linkers, including PEG, linear aliphatic chains, and more rigid (piperazine-type) linkers, have been studied, designing effective linkers remains challenging. Recently, DNA has been engineered to form DNA frameworks, such as DNA tetrahedra, octahedra, and icosahedra, with well-controlled surface chemistry. The rigidity, addressability, and artificial programmability of these DNA frameworks provide them with linker-like properties. Moreover, the caveolin-mediated endocytosis mechanism of DNA frameworks has been clarified, which is conducive to improving the poor cell entry efficiency of traditional PROTACs.

On July 27, 2023, Yi Ma, Department of Biomedical Engineering, School of Engineering, China Pharmaceutical University, China, and his team published a paper titled “DNA framework-engineered chimeras platform enables selectively targeted protein degradation” in Nature Communications. They have proposed DNA framework-engineered DbTACs with programmable linkers, that not only provides insight into general degrader design principles but also presents a promising strategy for guiding drug discovery.

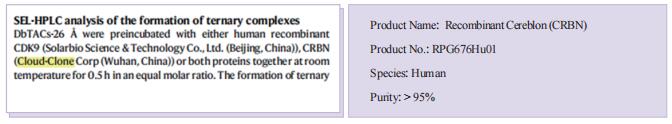
The protein [Recombinant Cereblon (CRBN), RPG676Hu01] of Cloud-Clone brand was chosed in this article, we are so proud for supporting the reaserchers.
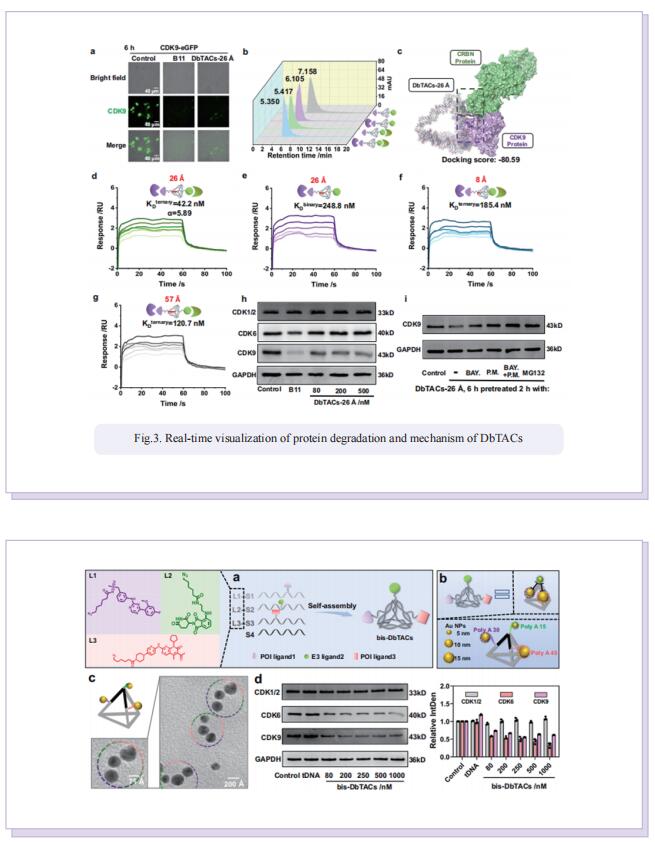
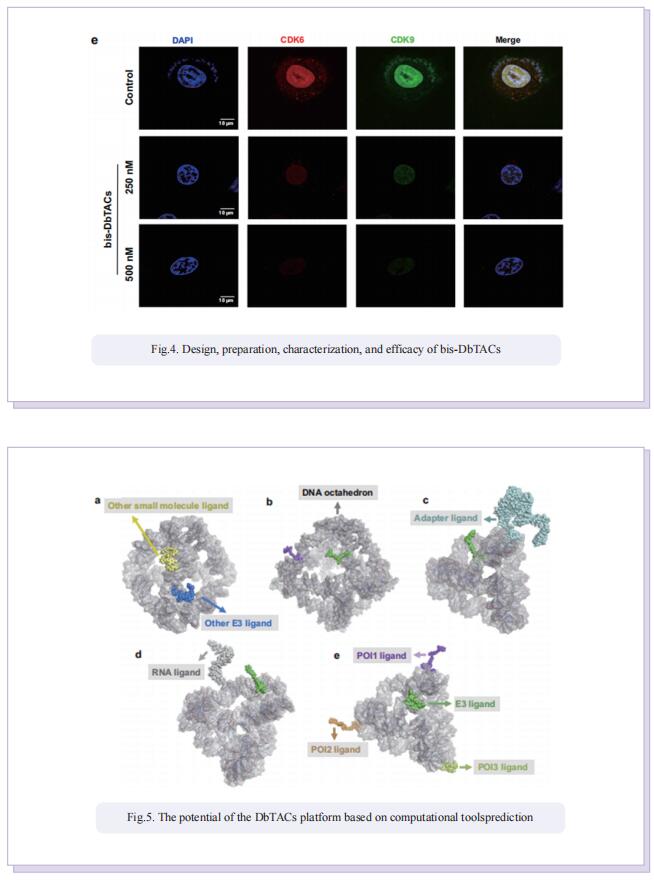
Inspired by the unique characteristics of DNA frameworks, the authors have developed an innovative strategy for the development of DNA framework-based PROTACs (DbTACs) by combining computational prediction, DNA self-assembly, and PROTACs technology. To study linker length-activity relationships, they employ DbTACs formed with DNA tetrahedra, the ligands of cyclin-dependent kinase (CDK) family protein and cereblon (CRBN) E3 ligase as representative templates. Specifically, the position of the CDK9 ligand is fixed, while the CRBN ligand is shuttled on DNA tetrahedra to produce DbTACs with linker lengths ranging from 8 to 57 Å. DbTACs with different linker lengths are visualized, and an optimal DNA linker length of 26 Å is found to be the most effective in vitro. The difference in binding affinity explains the mechanism of linker length–activity relationships. Furthermore, they successfully demonstrate the feasibility of their idea by creating bispecific DbTACs (bis-DbTACs) sharing one CRBN ligand, which selectively degrades CDK6 (a protein involves in cell proliferation and differentiation) and CDK9 (a protein that participates in transcriptional regulation, DNA repair, and metabolism) in cancer cell lines. To expand the scope of their platform, antibody-, and DNA motif-based DbTACs are also developed that facilitate the degradation of CDK9 kinase and ETS-related gene (ERG) transcription factor.
In summary, they have proposed DNA framework-engineered DbTACs with programmable linkers as an approach for the development of empirical nature in the linker domain. Compared to conventional PROTACs, DbTACs present a modular approach to generating degraders. This approach effectively reduces the number of steps involved in preparing the linker itself, as well as the subsequent coupling of the linker with the ligands. This represents a significant step forward in the development of more efficient drug production methods.
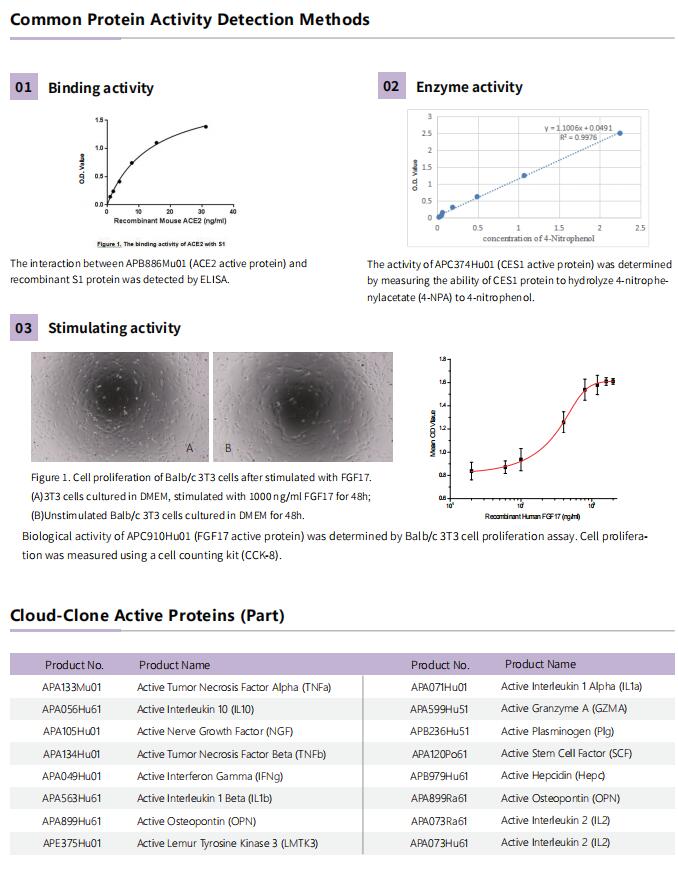
Cloud-Clone Active proteins based on GMP level development, can provide a variety of specifications of protein, meet the needs of basic science experiments, drug screening, animal drug delivery and other experiments.