New findings on the mechanism of lung cancer
Lung cancer is the leading cause of cancer-related death worldwide, with non-small cell lung cancer (NSCLC) being the most common type (~85%). NSCLCs are subdivided into lung adenocarcinomas (LUADs; ~50%), lung squamous cell carcinomas (LUSCs; ~30%), and others (~20%). Although disease understanding, treatment options, and outcomes for lung cancer are improving, survival continues to be low. 5-year survival in populations with lung cancer varies from 4-17% depending on stage and regional differences. Therefore, it is necessary to explore the underlying mechanisms of lung cancer development to support the design of new targeted therapeutic agents.
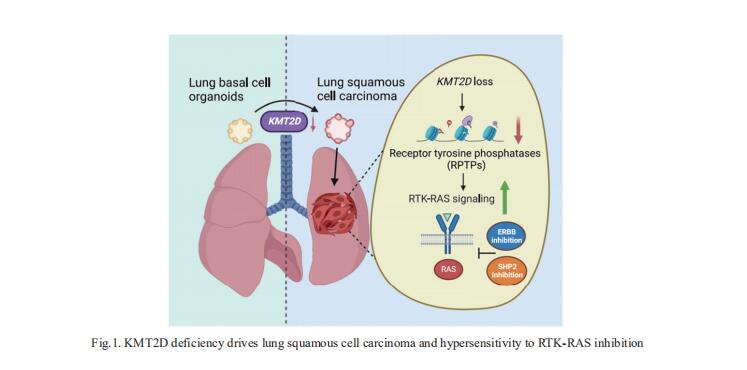
1. KMT2D deficiency drives lung squamous cell carcinoma and hypersensitivity to RTK-RAS inhibition
LUSC represents a major subtype of lung cancer with limited treatment options. KMT2D is one of the most frequently mutated genes in LUSC, and yet its role in LUSC oncogenesis remains unknown. Kwok-Kin Wong, Laura and Isaac Perlmutter Cancer Center, NYU Langone Health, USA, and his team identify KMT2D as a key regulator of LUSC tumorigenesis wherein Kmt2ddeletion transforms lung basal cell organoids to LUSC[1]. KMT2D loss increases activation of receptor tyrosinekinases (RTKs), EGFR and ERBB2, partly through reprogramming the chromatin landscape to repress the expression of protein tyrosine phosphatases. These events provoke a robust elevation in the oncogenic RTK-RAS signaling (Fig.1). Combining SHP2 inhibitor SHP099 and pan-ERBB inhibitor afatinib inhibits lung tumor growth in Kmt2d-deficient LUSC murine models and in patient-derived xenografts harboring KMT2D mutations. The study identifies KMT2D as a pivotal epigenetic modulator for LUSC oncogenesis and suggests that KMT2D loss renders LUSC therapeutically vulnerable to RTK-RAS inhibition.
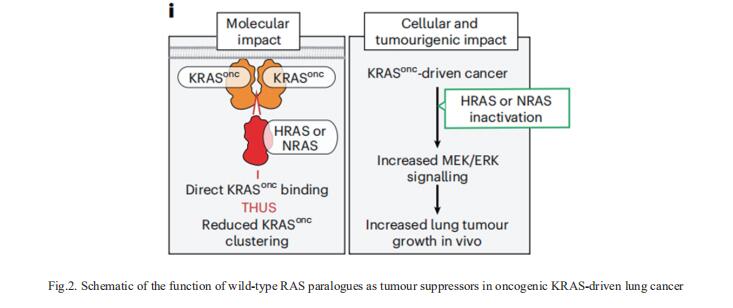
2. Multiplexed screens identify RAS paralogues HRAS and NRAS as suppressors of KRAS-driven lung cancer growth
Oncogenic KRAS mutations occur in approximately 30% of LUAD. Despite several decades of effort, oncogenic KRAS-driven lung cancer remains difficult to treat. Through a series of CRISPR/Cas9 screens in autochthonous lung cancer models, we show that HRAS and NRAS are suppressors of KRASG12D-driven tumour growth in vivo and confirm these effects in oncogenic KRAS-driven human lung cancer cell lines[2]. Mechanistically, RAS paralogues interact with oncogenic KRAS, suppress KRAS-KRAS interactions, and reduce downstream ERK signalling (Fig.2). Furthermore, HRAS and NRAS mutations identified in oncogenic KRAS-driven human tumours partially abolished this effect. By comparing the tumour-suppressive effects of HRAS and NRAS in oncogenic KRAS- and oncogenic BRAF-driven lung cancer models, they confirm that RAS paralogues are specific suppressors of KRAS-driven lung cancer in vivo. The study highlights the role RAS paralogue imbalance in oncogenic KRAS-driven lung cancer.
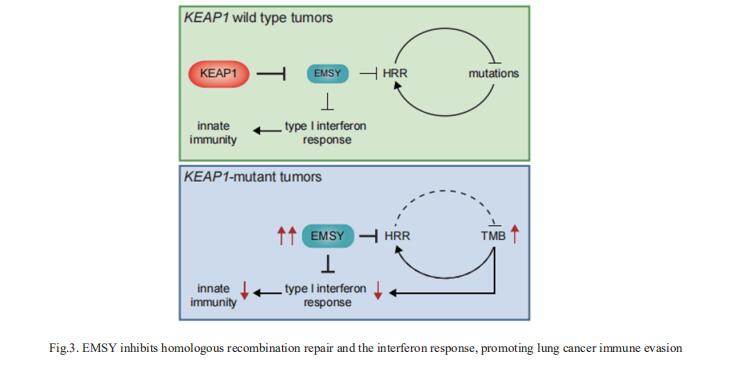
3. EMSY inhibits homologous recombination repair and the interferon response, promoting lung cancer immune evasion
NSCLCs harboring KEAP1 mutations are often resistant to immunotherapy. Michele Pagano, Department of Biochemistry and Molecular Pharmacology, New York University Grossman School of Medicine, USA, and his team showed that KEAP1 targets EMSY for ubiquitin-mediated degradation to regulate homologous recombination repair (HRR) and anti-tumor immunity[3]. Loss of KEAP1 in NSCLC induces stabilization of EMSY, producing a BRCAness phenotype, i.e., HRR defects and sensitivity to PARP inhibitors. Notably, EMSY accumulation suppresses the type I interferon response and impairs innate immune signaling, fostering cancer immune evasion (Fig.3). Activation of the type I interferon response in the tumor microenvironment using a STING agonist results in the engagement of innate and adaptive immune signaling and impairs the growth of KEAP1-mutant tumors. Their results suggest that targeting PARP and STING pathways, individually or in combination, represents a therapeutic strategy in NSCLC patients harboring alterations in KEAP1.
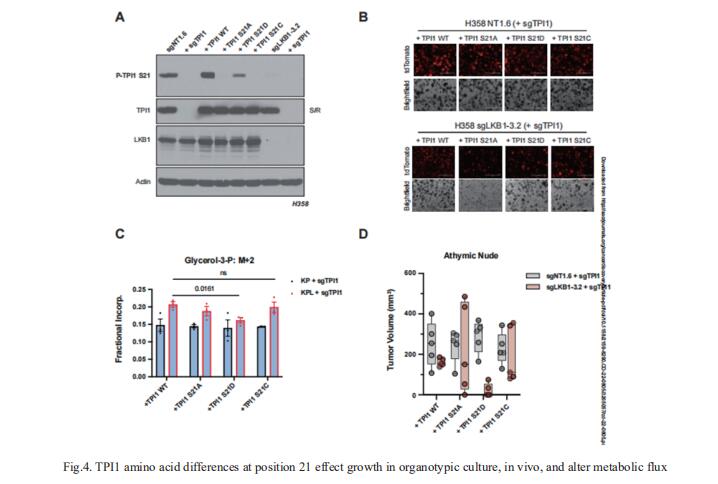
4. LKB1-dependent regulation of TPI1 creates a divergent metabolic liability between human and mouse lung adenocarcinoma
KRAS is the most frequently mutated oncogene in human lung adenocarcinomas (hLUAD) and activating mutations frequently co-occur with loss-of-function mutations in TP53 or STK11/LKB1. However, mutation of all three genes is rarely observed in hLUAD, even though engineered co-mutation is highly aggressive in mouse lung adenocarcinoma (mLUAD). Benjamin D. Stein, Weill Cornell Medicine, USA, and his team provided a mechanistic explanation for this difference by uncovering an evolutionary divergence in regulation of triosephosphate isomerase (TPI1)[4]. In hLUAD, TPI1 activity is regulated via phosphorylation at Ser21 by the Salt Inducible Kinases (SIKs) in an LKB1-dependent manner, modulating flux between completion of glycolysis and production of glycerol lipids (Fig.4). In mice, Ser21 of TPI1 is a Cys residue which can be oxidized to alter TPI1 activity without a need for SIKs or LKB1. The findings reported here suggest that selective inhibitors of LKB1 or of SIK family protein kinases could be effective in treating human KRAS/TP53 mutant lung cancers or other cancers with KRAS/TP53 mutations.
References
[1]Pan Y, Han H, Hu H, et al. KMT2D deficiency drives lung squamous cell carcinoma and hypersensitivity to RTK-RAS inhibition [J]. Cancer Cell. 2023;41(1):88-105.e8. (IF=38.585)
[2]Tang R, Shuldiner EG, Kelly M, et al. Multiplexed screens identify RAS paralogues HRAS and NRAS as suppressors of KRAS-driven lung cancer growth [J]. Nat Cell Biol. 2023;25(1):159-169. (IF=28.213)
[3]Marzio A, Kurz E, Sahni JM, et al. EMSY inhibits homologous recombination repair and the interferon response, promoting lung cancer immune evasion [J]. Cell. 2022;185(1):169-183.e19. (IF=66.850)
[4]Stein BD, Ferrarone JR, Gardner EE, et al. LKB1-dependent regulation of TPI1 creates a divergent metabolic liability between human and mouse lung adenocarcinoma [J]. Cancer Discov. 2023;CD-22-0805. (IF=38.272)
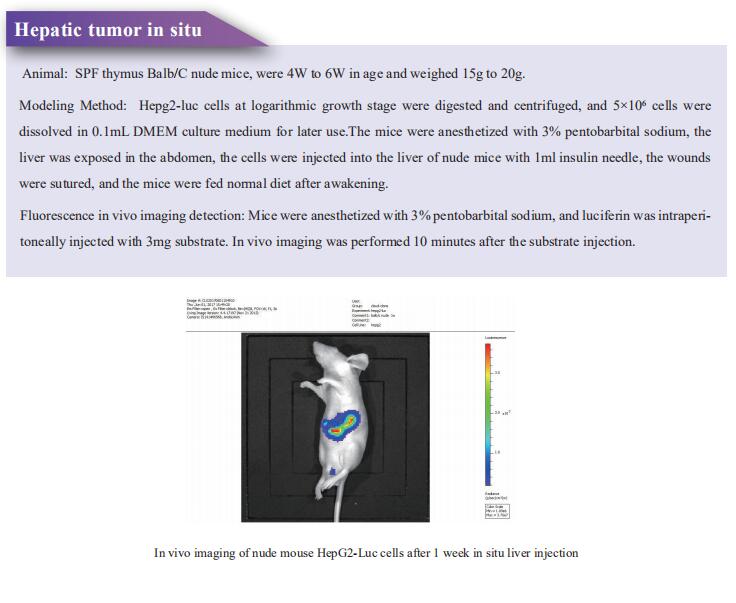
Cloud-Clone not only provides a variety of tumor experimental animal models, including tumor transplantation animal models, spontaneous tumor animal models, induced tumor animal models, tumor metastasis animal models, etc., covering common tumor research. We also have various cancer detection indicators and the above-mentioned EGFR, ERBB2, HRAS, NRAS, KRAS, BRAF, TPI1, SIK, LKB1 and other related products, which can help scientific researchers to conduct cancer-related research.